Evo: DNA foundation modeling from molecular to genome scale

Introducing Evo
Evo is a long-context biological foundation model that generalizes across the fundamental languages of biology: DNA, RNA, and proteins. It is capable of both prediction tasks and generative design, from molecular to whole genome scale (over 650k tokens in length). Evo is trained at a single-nucleotide (byte) resolution, on a large corpus of prokaryotic and phage genomic sequences.
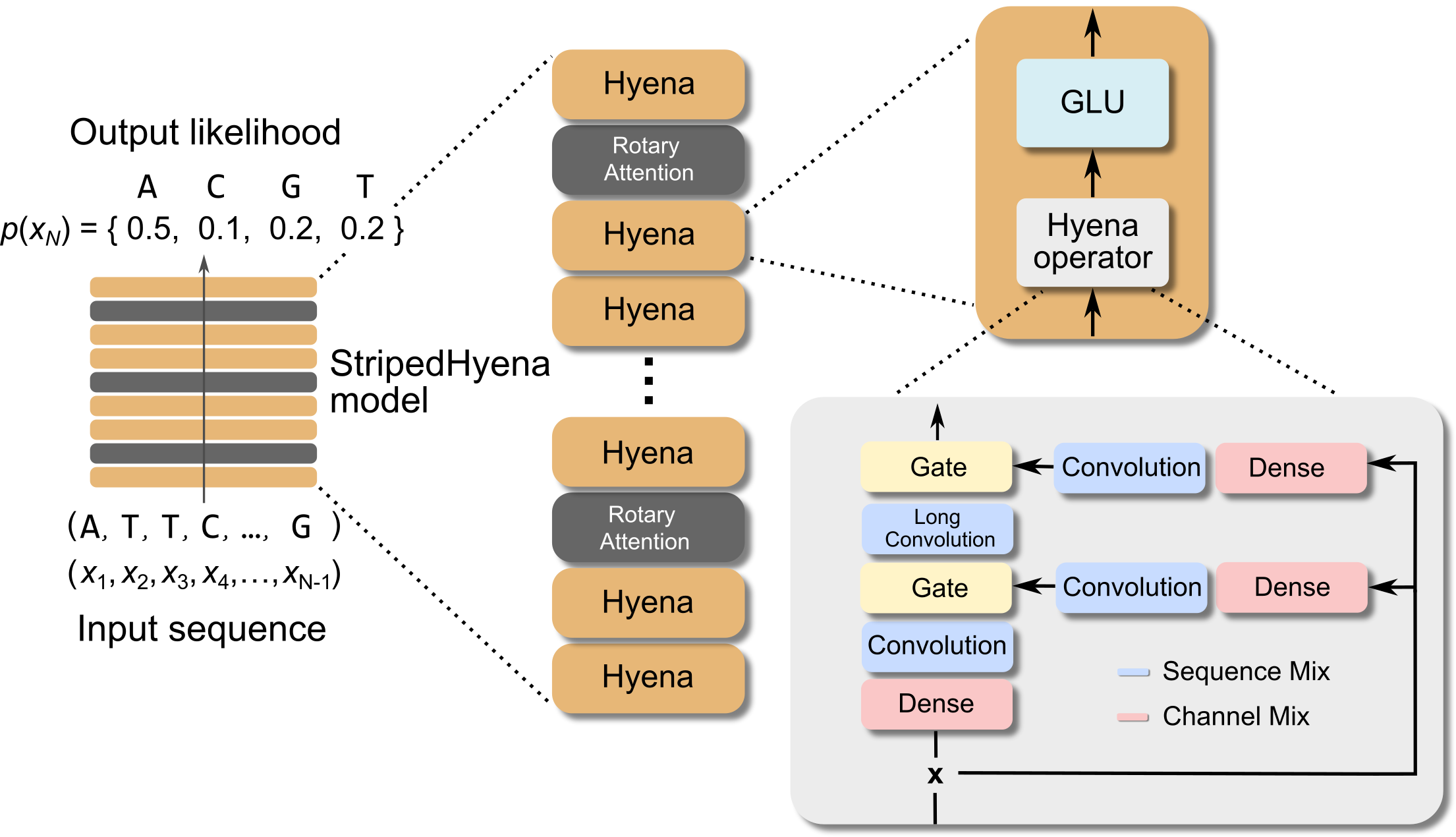
Evo is a 7 billion parameter model trained to generate DNA sequences using a context length of 131k tokens, and is based on StripedHyena, a deep signal processing architecture designed to improve efficiency and quality over the prevailing Transformer architecture. Evo was developed by Arc Institute, Stanford, and TogetherAI researchers.
Evo is available via our GitHub repository and the Together playground, where you can generate DNA in your browser. In addition to model weights, we are excited to release intermediate checkpoints on HuggingFace. We're also open-sourcing a large 300B token training dataset we compiled, which we call OpenGenome, consisting of 2.7M publicly available prokaryotic and phage genomes, in the coming days. It's the largest DNA pretraining dataset publicly available, which we hope will help accelerate research in this exciting and impactful domain of DNA language modeling.
Is DNA all you need?
In biology, everything starts with DNA. Genomes carry an entire set of DNA (the genetic code) to make a complete organism. Within them lies the result of generations of evolution, reflecting adaptations to constantly shifting environmental changes. Other complex biological languages emerge from this code, including proteins, the tiny molecular machines that make cells function, and RNA, which helps DNA transmit information and often helps proteins accomplish their functions. As multilayered as these languages seem, they are all unified in (our) genomes.
The emergence of AI foundation models has charted a promising path in biological sequence modeling, yet modeling at the whole genome level has been out of reach for many methods. DNA sequences are extremely long (up to billions of nucleotides), and the sensitivity required to fully understand the effects of evolution (which occurs one nucleotide at a time), makes it a particularly challenging domain for large-scale pretraining. So far, it’s been unclear if AI models are able to effectively learn such complex patterns over these long ranges. As a result, existing breakthroughs in modeling biological sequences with AI have instead focused on short context, task-specific, and single-modality capabilities (e.g. AlphaFold, ESMFold, Nucleotide Transformer).
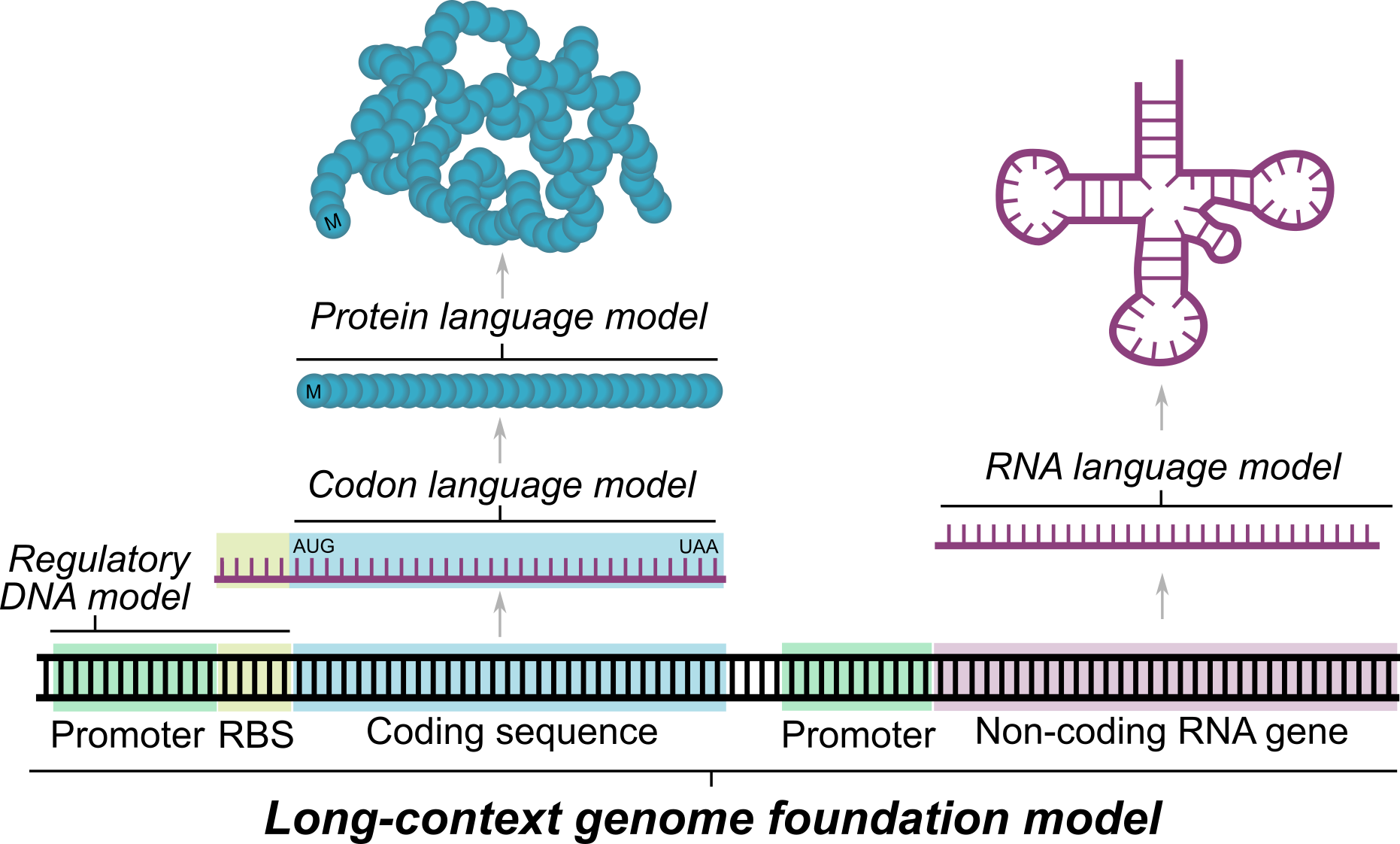
Evo is a DNA foundation model
These challenges (and the fundamental question of whether DNA is all you need) motivated us to work on Evo. In particular, we wanted a foundation model that could integrate information over long genomic sequences while retaining sensitivity to single-nucleotide changes. A model that effectively learns over genomes could understand not only the individual DNA, RNA, and protein components, but also how these interact to create complex systems. This could accelerate our mechanistic understanding of biology and the ability to engineer life itself.
Demonstrating the first scaling laws on DNA pretraining
To overcome the challenges associated with sequence modeling at long sequence lengths and at byte-level resolution, we used the newly developed StripedHyena architecture, which achieves both long context and nucleotide resolution via our latest advances in architecture design. StripedHyena works by hybridizing rotary attention and hyena operators to efficiently process and recall patterns in long sequences.
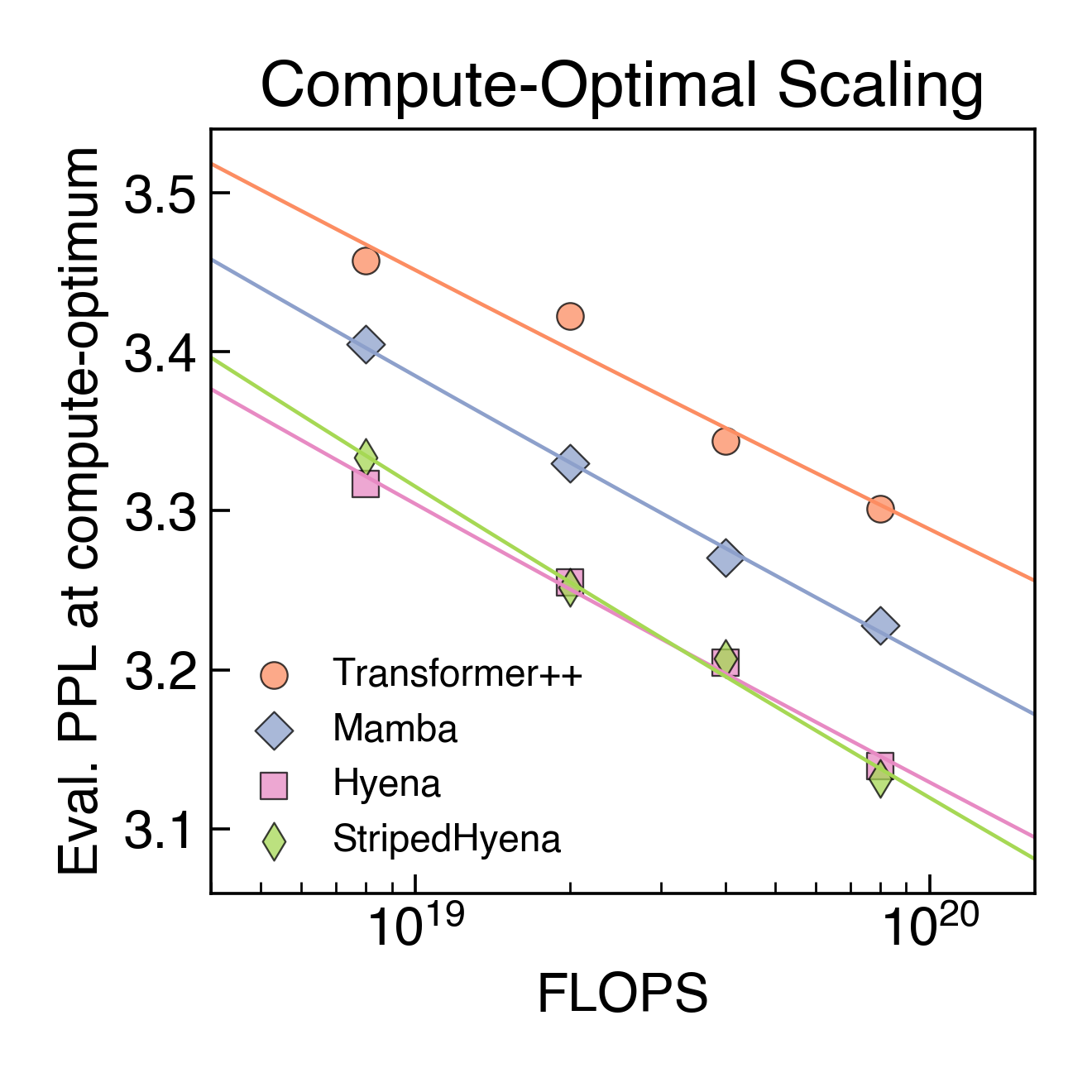
To aid our model design and optimize performance at scale, we performed the first scaling laws analysis on DNA pretraining across leading architectures (Transformer++, Mamba, Hyena, and StripedHyena), training over 300 models from 6M to 1B parameters at increasing compute budgets (FLOPS). We saw that Transformer models do not scale as well when trained at a single-nucleotide, byte-level resolution, indicating that the predominant architecture in natural language doesn’t necessarily transfer to DNA.
Evo capabilities
Zero-shot gene essentiality testing
Strikingly, Evo understands biological function at the whole genome level. Using an in silico gene essentiality test, Evo can predict which genes are essential to an organism’s survival based on small DNA mutations. It can do so zero-shot and with no supervision. For comparison, a gene essentiality experiment in the laboratory could require 6 months to a year of experimental effort. In contrast, we replace this with a few forward passes through a neural network.
Zero-shot prediction across DNA, RNA, and protein modalities
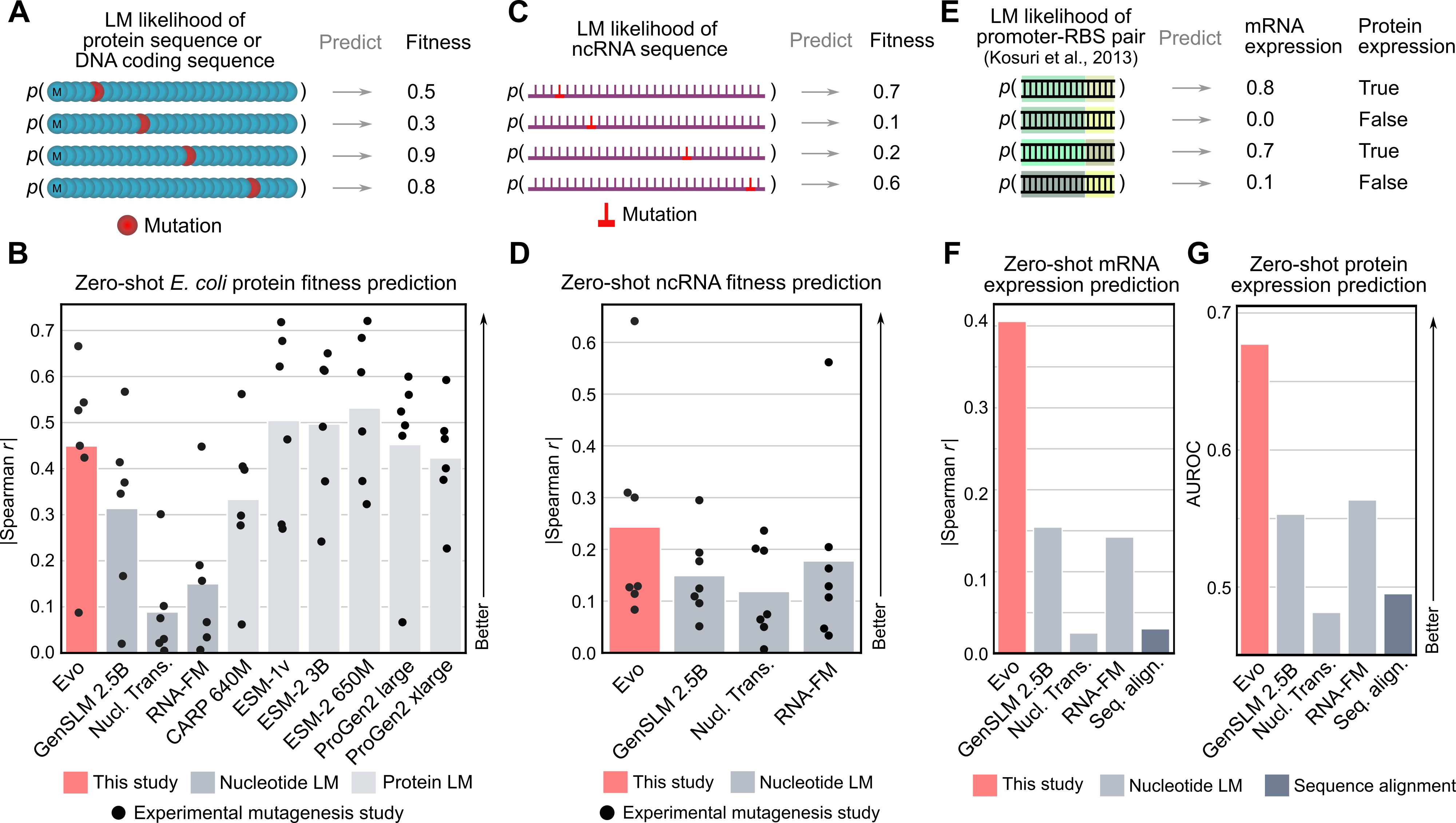
Because Evo is trained on long genomic sequences that contain protein coding sequences, we tested whether the model would also learn the protein language well enough to perform zero-shot protein function prediction. Evo outperforms all other nucleotide models tested, including models explicitly trained only on protein coding sequences, and is even competitive with state-of-the-art protein language models, like ESM or ProGen. But there are more than just proteins in Evo’s genomic training data—there are ncRNAs and regulatory DNA sequences in genomes as well. Notably, we show that Evo enables zero-shot function prediction for ncRNA and regulatory DNA, as well, thereby spanning all three modalities of the central dogma.
CRISPR system generation
Today, generative models for biology usually focus on a single modality (at a time)—for example, only on proteins or on RNA. One of the key breakthroughs we highlight is that Evo can perform multimodal design to generate novel CRISPR systems, a task that requires creating large functional complexes of proteins and ncRNA (non-coding RNA), and is out of reach for existing generative models. Typically, discovering new CRISPR systems requires searching through natural genomes for similar sequences that were literally taken from an organism. Instead, Evo enables a new approach to generating biological diversity by sampling sequences directly from a generative model, an exciting frontier for creating new forms of genome editing tools.
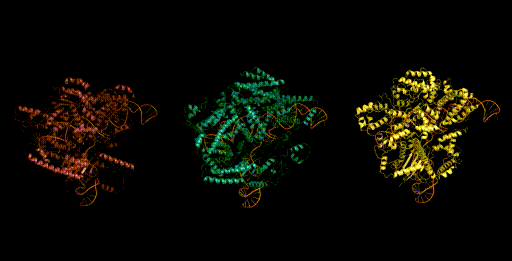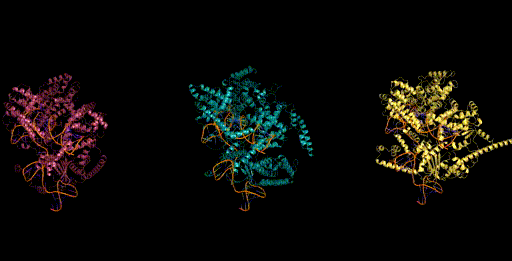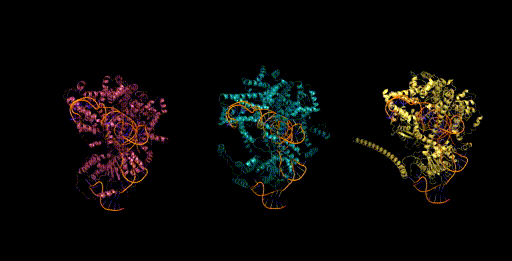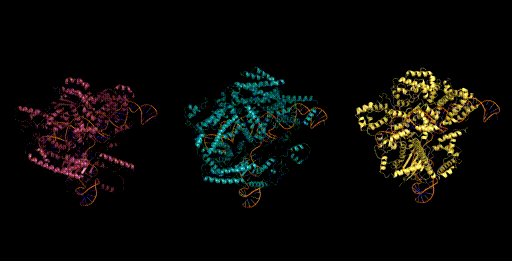
Genome scale generation
In addition to generating at the scale of multiple molecules (proteins and ncRNA), Evo has the potential to generate sequences at the scale of whole genomes. Generating sequences at this scale is enabled by both the long context capabilities of the architecture, as well as from its efficient inference mode. We generated sequences of over 650k nucleotides on a single GPU. When we sample sequences at this length with Evo, we find genomes that contain thousands of potential protein-coding sequences.
Safe and responsible development of Evo
Evo is the first of its kind to predict and generate DNA sequences at the whole genome scale with single-nucleotide resolution. Future capabilities that emerge from large-scale DNA models like Evo also require additional work to ensure that these capabilities are deployed safely and for the benefit of humanity. In our paper, we provide an extended discussion on potential risks and precautionary measures.
Future plans
Evo marks a turning point in what we think is possible in modeling biological sequences, and beyond. We believe this technology has the potential to accelerate discovery and understanding in the sciences (such as biology, chemistry, or material science), as well as be applied to real-world problems including drug discovery, agriculture, and sustainability. Although the results show promising computational capabilities, further experimental validation is required for the generated sequences.
Foundation models are going to be increasingly important scientific tools. We look forward to training larger models, improving their generation capabilities, and expanding Evo pretraining to human genomes. By enhancing the biological complexity learned by these models, we believe we can make significant progress toward fighting complex diseases and improving human health.
Acknowledgments
Brian Hie
Patrick Hsu
Eric Nguyen
Michael Poli
Matthew Durrant
This work was only possible with an incredibly talented group of researchers. The full team also includes: Armin Thomas, Brian Kang, Jeremy Sullivan, Madelena Ng, Ashley Lewis, Aman Patel, Aarou Lou, Stefano Ermon, Stephen Baccus, Tina Hernandez-Boussard, and Chris Ré.
Resources
Manuscript (Now published in Science doi: https://doi.org/10.1126/science.ado9336)
Together playground (generate DNA in browser)
Code
HuggingFace checkpoint
pip install
Hazy Research
Hsu Lab
Hie Lab
Arc Tools Portal





