Introducing the Arc Tool Portal
As the life sciences become increasingly interdisciplinary and computational, the application of machine learning to biological problems has enabled a new revolution in predictive models that can make sense of large, complex biological datasets.
At Arc, we're investing heavily in the infrastructure and expertise required to push forward the frontier of machine learning in biology across our faculty and Technology Centers. Part of our vision to advance the use of machine learning in biomedical research is to develop and disseminate computational resources and tools that enable reproducibility and support discovery throughout the scientific community.
The Arc Tools Portal will be an ever-growing one-stop shop for computational tools, developed and maintained by scientists at Arc. In the future, we will expand it to include protocols, datasets, open-source software, and other resources that can help researchers across the scientific community to fast track their own research.
Using deep learning to target RNA with high precision and efficacy
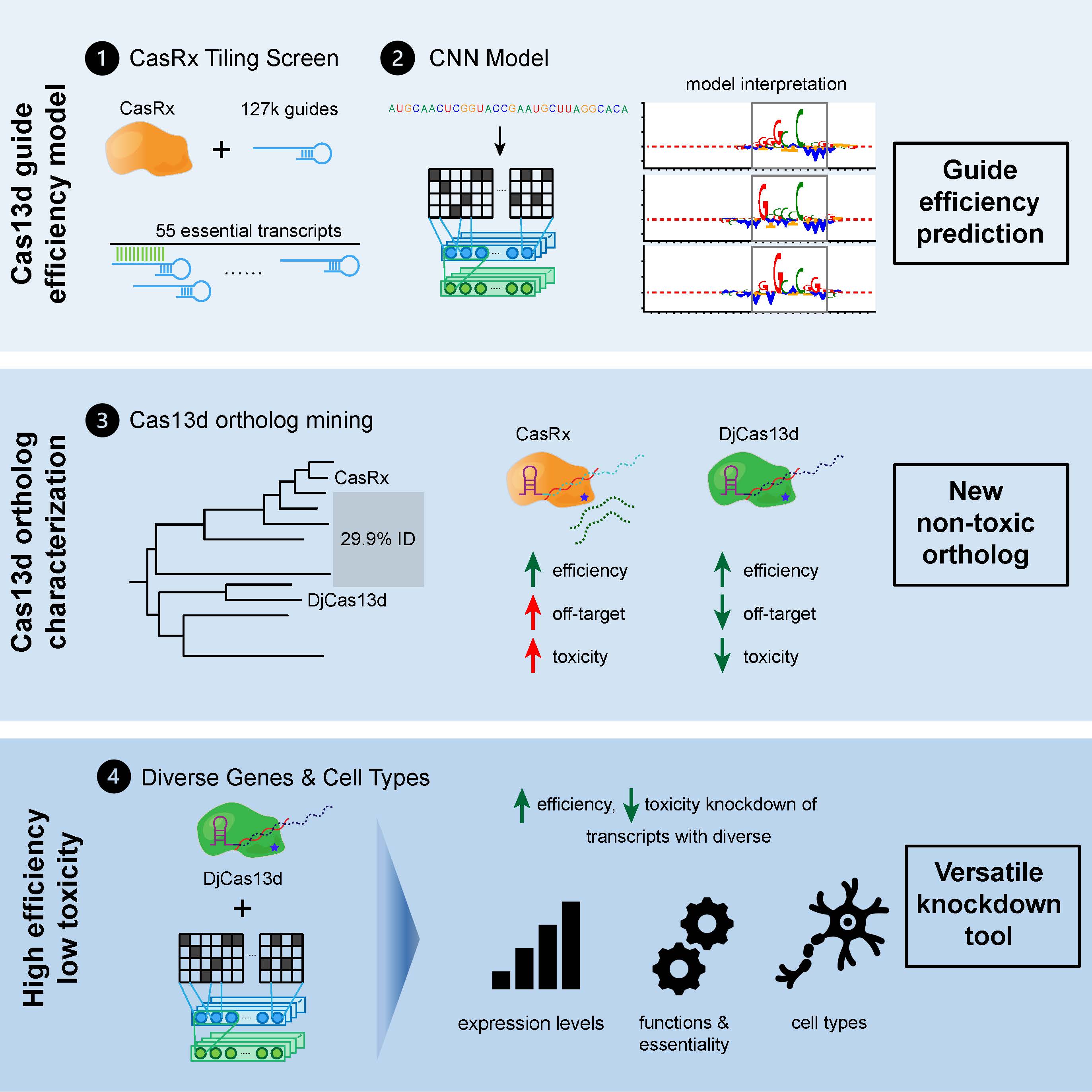
One of our first tools released today is based on a new paper from the Konermann and Hsu labs at Arc – led by Stanford Bioengineering Ph.D student Jingyi Wei. The paper, published today in Cell Systems, reports the development of a convolutional neural network (CNN) model that predicts highly efficient guide RNA sequences for CRISPR-Cas13d-based transcriptome engineering. What’s more, the neural network is not simply a “black box”: it sits at the frontier of deep learning models that now enables researchers to use algorithms to interpret how a model makes decisions, and learn mechanistic principles for future R&D.
While CRISPR-based genome engineering systems such as Cas9 have revolutionized our ability to target DNA for discovery research and therapeutic applications, our ability to target RNAs in the cell in an efficient, precise and easily reprogrammable way has been lagging behind. RNA is the continuously changing counterpart of DNA, controlling cellular and organism function during both health and disease, but its output from the DNA genome varies highly between different cell types and cell states. Molecular and computational tools to target RNA would therefore be highly impactful to manipulate these dynamic processes.
In prior work, Konermann and colleagues described the first single-enzyme, RNA-targeting CRISPR system in vitro – now termed Cas13a. This biochemical work unraveled the unique targeting mechanism of RNA-targeting CRISPR systems and laid the groundwork for engineering of these systems for applications in human cells. A follow-on study by Hsu and Konermann described the discovery and engineering of a related but unusually small and efficient family of such RNA targeting systems, Cas13d.
CRISPR-Cas13d is a programmable tool that allows researchers to find and destroy the RNA copies of a particular gene of interest in human cells. As a counterpart to DNA nuclease tools, RNA targeting with Cas13d has the advantage of being able to transiently and gradually modulate gene expression without any permanent alterations to the DNA. It can also be adapted to target RNAs for many other applications such as imaging, alteration of splicing, or targeted chemical modifications.
While the initial – previously published – variant of Cas13d was often highly effective across these applications, two major limitations remained.
The first limitation is shared across essentially all genome engineering systems: when trying to target a particular RNA transcript (or DNA sequence), not all guide sequences will work equally well. “Currently, a researcher using Cas13d for RNA targeting would have to manually test on average 3-4 separate guide RNAs to find a highly potent one,” said Wei. “We were able to overcome the need for this by performing a large scale screen of more than 130,000 different guide RNAs in combination with a deep learning model. This enabled us to unravel the rules that determine which guide RNAs will be most effective.” Indeed, the tool was able to accurately choose nine highly effective guides out of 10 selected, across a variety of contexts. This is in comparison to a baseline of two out of ten when not using the tool – an impressive performance that significantly outperformed other published tools.
Besides variable efficacy, the second limitation of the earlier generation of all Cas13-based RNA targeting CRISPR systems was the potential for generating undesired collateral damage in specific contexts. This could lead to problems especially when targeting RNAs in sensitive cell types. To overcome this challenge, Wei and colleagues took a computational approach again – this time mining the natural diversity of Cas13d enzymes across microbiome datasets. This search resulted in an almost 10-fold expansion of the known diversity of Cas13d systems, and was followed by comprehensive screening of more than 50 enzyme variants, revealing a novel enzyme – termed DjCas13d – from an Australian sheep microbiome assembly that has a much higher degree of specificity.
“Taken together, the study and tool released today allow for precise and efficient RNA targeting even in highly sensitive human cell types – including human embryonic stem cells, neurons and hematopoietic progenitor cells – representing a major improvement of transcriptome engineering technology,” said Konermann. “This new study is also a great example of the types of tools we’re excited about at Arc: innovations that integrate big data with deep subject-matter expertise and cutting-edge machine learning that can overcome major challenges in biotechnology.”
Building out the Arc Tools Portal
Looking ahead, we’re eager to grow our tools hub into a highly useful resource for the scientific community. The second tool we released today is just the beginning of this expansion.
ScreenPro2, which was developed by Abe Arab, a Research Assistant in the Gilbert lab in collaboration with our Multi-Omics Technology Center, is an accessible, Python-based toolbox for end-to-end analysis of data from CRISPR screens. In particular, Screenpro2 enables users who are interested in pooled CRISPR screens to easily analyze experiments performed with standard or custom single and dual sgRNA library designs.
“At Arc, we want to make resources, including analytical approaches, available so that they can benefit the broader research community and advance scientific progress,” said Luke Gilbert, Arc Core Investigator and Associate Professor at UCSF. “Screenpro2 is a perfect example of how we’re putting this into practice. The tool simplifies analysis of cutting edge CRISPR libraries, thus enabling greater use by the broader scientific community.”
Computational tool development is only one part of the major technology development efforts underway at Arc. Across Arc, our scientists are hard at work developing cutting-edge techniques in genome engineering, multi-omics and advanced cellular models – just to name a few. Along with the computational and analytical components of these efforts, we will also make in-depth experimental protocols, reagents and datasets available on the Tools Portal in the future, so that other labs can adopt our emerging technologies and build on them.
