
Engineered CRISPR-Cas12a for higher-order combinatorial chromatin perturbations
C. C.-S. Hsiung, C. M. Wilson, N. A. Sambold, R. Dai, Q. Chen, N. Teyssier, S. Misiukiewicz, A. Arab, T. O'Loughlin, J. C. Cofsky, J. Shi & L. A. Gilbert

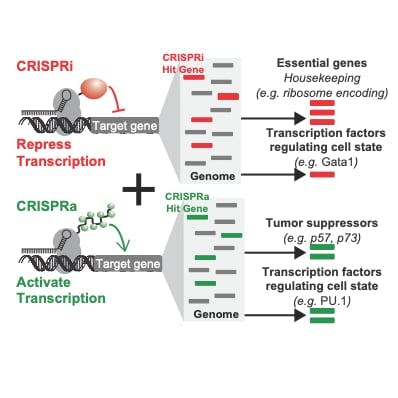
We have pioneered the use of a nuclease inactivated version of CRISPR/dCas9 in human cells to edit the epigenome or control transcription of protein coding or non-coding genes. We use dCas9 as a general RNA-guided DNA binding platform to recruit enzyme activities or protein complexes to specific target sites in the genome to execute a desired program. We have shown we can turn on (CRISPRa) and off (CRISPRi) expression of most genes encoded by the human genome. These tools allow us to mechanistically determine how epigenetic or transcriptional states determine cancer cell behavior and response to anti-cancer interventions. We will continue to use synthetic biology to build new innovative tools for editing the epigenome!
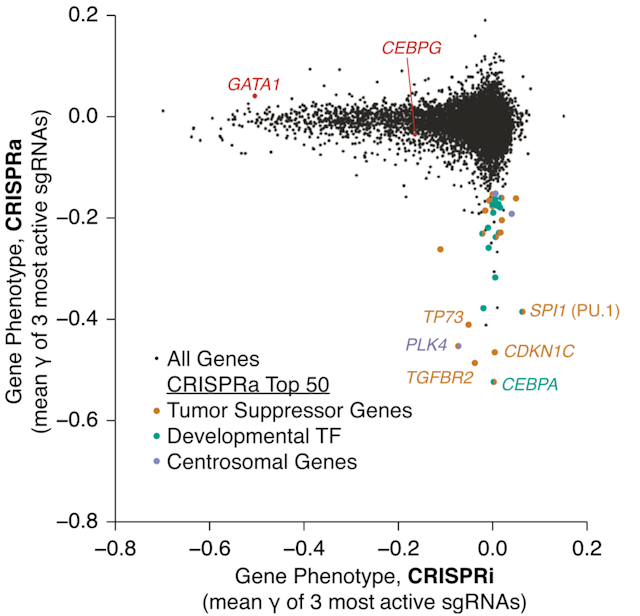
We have developed genome scale CRISPRi/a sgRNA libraries targeting protein coding and non-coding genes encoded by the human and mouse genome. We are using CRISPRi and CRISPRa screens to elucidate genes that dictate response to anti-cancer drugs or determine cancer cell properties such as metastasis or developmental plasticity. We have shown that genome scale CRISPRi/a screens reveal complementary principles of oncogene and non-oncogene addictions.
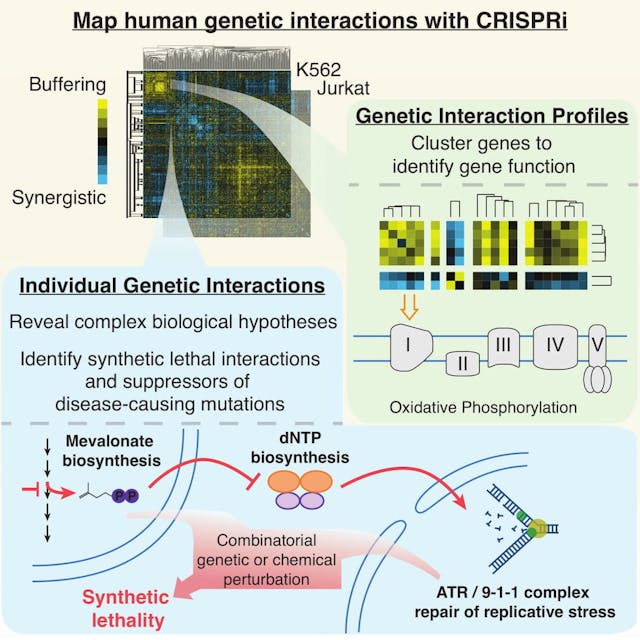
Genetic interaction maps are pairwise perturbations that measure gene epistasis relationships. We are building genetic interaction maps to search for synthetic lethal gene relationships in cancer, map signaling networks and determine the functions of poorly characterized genes encoded by the human genome.

C. C.-S. Hsiung, C. M. Wilson, N. A. Sambold, R. Dai, Q. Chen, N. Teyssier, S. Misiukiewicz, A. Arab, T. O'Loughlin, J. C. Cofsky, J. Shi & L. A. Gilbert

James K Nuñez, Jin Chen, Greg C Pommier, J Zachery Cogan, Joseph M Replogle, Carmen Adriaens, Gokul N Ramadoss, Quanming Shi, King L Hung, Avi J Samelson, Angela N Pogson, James Y S Kim, Amanda Chung, Manuel D Leonetti, Howard Y Chang, Martin Kampmann, Bradley E Bernstein, Volker Hovestadt, Luke A Gilbert, Jonathan S Weissman

Max A Horlbeck, Albert Xu, Min Wang, Neal K Bennett, Chong Y Park, Derek Bogdanoff, Britt Adamson, Eric D Chow, Martin Kampmann, Tim R Peterson, Ken Nakamura, Michael A Fischbach, Jonathan S Weissman, Luke A Gilbert

Luke Gilbert is an Associate Professor of Urology and of Cellular and Molecular Pharmacology at the University of California, San Francisco, and is affiliated with the Helen Diller Family Comprehensive Cancer Center and the Innovative Genomics Institute. He received his Ph.D. from MIT and completed his postdoctoral training at UCSF. Dr. Gilbert was an early pioneer in applying CRISPR technology to edit the epigenome (CRISPRi and CRISPRa), and continues to develop new strategies for editing and understanding human genetics and epigenetics at large scale, with a particular focus on cancer. Dr. Gilbert’s research has been recognized by a NIH Director’s New Innovator Award, a Pew-Stewart scholar for cancer research award, and the AAAS Martin and Rose Wachtel Cancer research award.

Columbia University (Biochem MD/Ph.D.)

University of Pennsylvania (M.D., Ph.D.)

Western New England University (Forensic Sciences, B.A. & Biology, B.A.)

Lewis and Clark College (Biochemistry, B.A.)

University of Richmond (Biology, Computer Science B.S.)

Scripps College (Biochemistry B.A.)

Northwestern University (Biology & Economics B.A.)

Reed College (Biochemistry B.A.)

Colby College (Biology B.A.)

UC Berkeley (MCB, CS B.A.)

Sonoma State University (B.ASc. Biology)

Dickinson College (Mathematics, Computer Science B.S.)

UCSF (PhD)

McGill University (B.Eng Bioengineering)




